File format converter¶
Overview¶
File format converter will realize Neural Network Libraries (or Console) workflow with ONNX file format, and also NNabla C Runtime.
File format converter has following functions.
Convert NNP variations to valid NNP
Convert ONNX to NNP
Convert NNP to ONNX
Convert NNP to NNB(Binary format for NNabla C Runtime)
Convert NNP to Tensorflow saved_model
Convert Tensorflow checkpoint, frozen graph or saved_model to NNP
Convert NNP to Tensorflow Lite
Convert Tensorflow Lite to NNP
Experimental: Convert NNP to C Source code for NNabla C Runtime
IMPORTANT NOTICE: This file format converter still has some known problems.
Supported ONNX operator is limited. See Function-Level Support Status.
Supported Tensorflow operator is limited. See Function-Level Support Status.
Converting NNP to C Source code is still experimental. It should work but did not tested well.
Architecture¶
This file format converter uses protobuf defined in Neural Network Libraries as intermediate format.
While this is not a generic file format converter, this is the specified converter for Neural Network Libraries.
This converter can specify both inputs and outputs for ONNX file, but if ONNX file contains a function unsupported by Neural Network Libraries, it may cause error in conversion.
This converter also provides some intermediate process functionalities. See Process.
Installation¶
Before using this converter, please use command pip install nnabla_converter to install nnabla_converter.
Note that, flatbuffer package is necessary for TFLite export, please check Tensorflow Lite section in this page for more details.
Conversion¶
Supported Formats¶
NNP¶
NNP is file format of NNabla.
NNP format is described at Data Format.
But with this file format converter is work with several variation of NNP.
Standard NNP format (.nnp)
Contents of NNP files(.nntxt, .prototxt, .h5, .protobuf)
ONNX¶
Limitation¶
Training is not supported.
Support operator set 7,9,10,11.
Not all functions are supported. See Function-Level Support Status.
Only limited Neural Network Console projects supported. See Model Support Status.
NNB¶
NNB is compact binary format for NNabla C Runtime. The file format is shown as the following diagram:
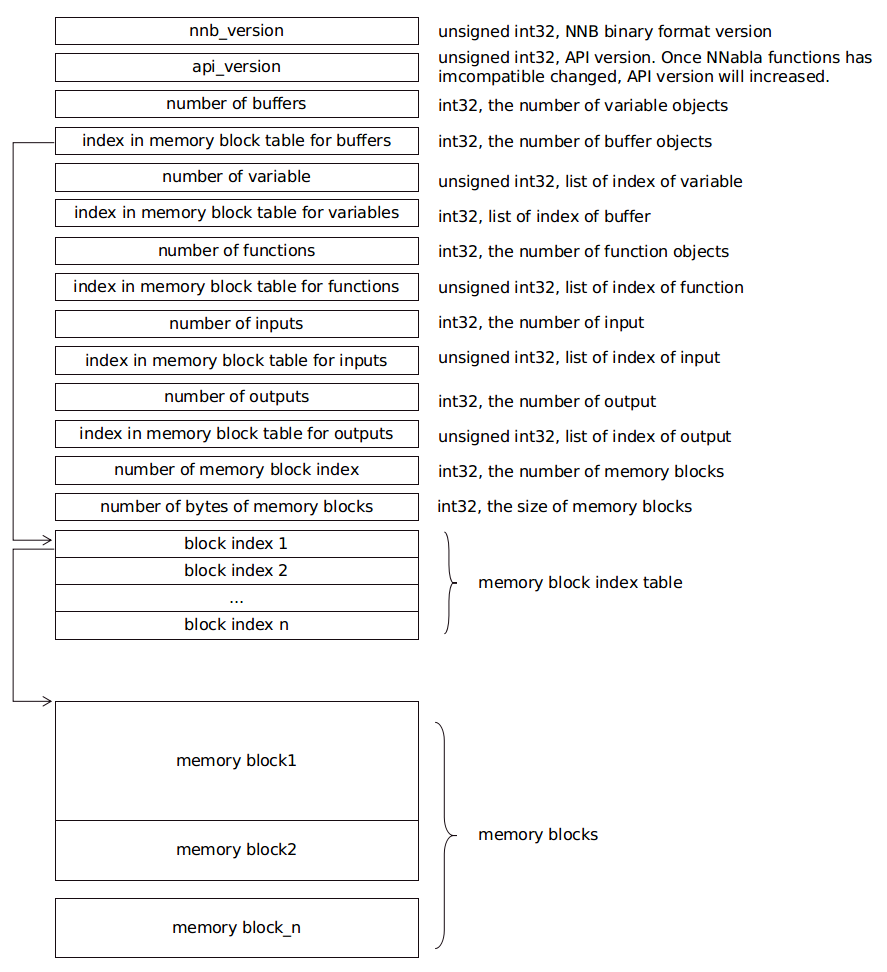
There are several concepts, such as buffer, variable, function, input and output in this file. Each of them is represented as a list. Each list is recorded with 2 members: number of object, and index in memory block table. The index points to the position in a memory block index table. The index in memory block index table points to the start address of memory data block.
It is designed for nnabla-c-runtime.
C Source Code¶
File format converter supports C source code output for nnabla-c-runtime.
Tensorflow¶
Limitation¶
Bridged by onnx, tensorflow import and export is supported with some limitations.
- As for the importer, 4 formats tends to be supported:
.pb, tensorflow frozen graph format
.ckpt, tensorflow check point format version 1
.ckpt.*, tensorflow check point format version 2
saved_model, tensorflow saved_model format
As for the exporter, some of Neural Network Console projects are supported. See Model Support Status. The output of converter is tensorflow saved_model format.
Tensorflow Lite¶
Limitation¶
- For export to tensorflow lite, please install
flatbufferspackage: For Windows platform, download package from FlatBuffers and extract.
For Linux platform, use command
snap install flatbuffersto install flatbuffers.For MaxOS platform, use command
brew install flatbuffersto install flatbuffers.
and add the executable file flatc to the system PATH.
After exporting TFLite, a json file with the same name will be generated, recording whether the input and output of the TFLite network need to be transposed to channel_last according to base_axis.
Process¶
Expand Repeat and Recurrent¶
Neural Network Console supports LoopControl pseudo functions RepeatStart, RepeatEnd, RecurrentInput, RecurrentOutput or Delay.
Currently, these functions are not supported by Neural Network Libraries directly.
The file format converter expands the network and removes these pseudo functions by default.
If you want to preserve these, specify command line option --nnp-no-expand-network when converting files.
Split network¶
You can split network with --split option.
See Splitting network to use this functionality.
Usage¶
NNP Operation¶
Convert NNP to NNP¶
Sometimes we need to convert NNP to NNP.
Most major usecase, expand repeat or recurrent network supported by Neural Network Console but not supported by C++ API.
$ nnabla_cli convert input.nnp output.nnp
Convert console output to single NNP file¶
Current version of Neural Network Console outputs .nntxt and .h5 as training result.
Then we need to convert separated files into single NNP and parameters store with protobuf format.
$ nnabla_cli convert net.nntxt parameters.h5 output.nnp
Convert console output to single NNP file without expanding Repeat or recurrent.¶
$ nnabla_cli convert --nnp-no-expand-network net.nntxt parameters.h5 output.nnp
Keep parameter format as hdf5¶
$ nnabla_cli convert --nnp-no-expand-network --nnp-parameter-h5 net.nntxt parameters.h5 output.nnp
Everything into single nntxt.¶
$ nnabla_cli convert --nnp-parameter-nntxt net.nntxt parameters.h5 output.nntxt
ONNX Operation¶
Convert NNP to ONNX¶
$ nnabla_cli convert input.nnp output.onnx
If specify output onnx opset 9, please use the following (default is opset 7):
$ nnabla_cli convert input.nnp output.onnx -d opset_9
Convert ONNX to NNP¶
$ nnabla_cli convert input.onnx output.nnp
Currently, opset 7,9,10,11 are supported to import.
C Runtime Operation¶
Generally, it is better to set the batch size to 1 when convert file to C runtime.
If the batch size is larger than 1, it is necessary to process the batch size data collectively
To make the batch size 1, add -b 1 to command line option.
Convert NNP to NNB¶
$ nnabla_cli convert -b 1 input.nnp output.nnb
Convert NNP to C source code¶
$ nnabla_cli convert -b 1 -O CSRC input.onnx output-dir
Quantization¶
C-runtime library supports binary(or fixed point) weights, which can dramatically downsize the model (and footprint). See Compress network by fixed point quantization for how to quantize your model.
Tensorflow Operation¶
Convert NNP to Tensorflow saved_model¶
$ nnabla_cli convert input.nnp output_saved_model --export-format SAVED_MODEL
Convert NNP to Tensorflow frozen graph¶
$ nnabla_cli convert input.nnp output.pb
Convert Tensorflow frozen graph to NNP¶
$ nnabla_cli convert input.pb output.nnp
Convert Tensorflow checkpoint to NNP¶
For checkpoint version 1:
$ nnabla_cli convert input.ckpt output.nnp --inputs x0,x1 --outputs y0,y1
In the same directory of input.ckpt, the related files, such as checkpoint, input.ckpt.meta and so on are required
to exist. The inputs required the input name of model, separated by comma. The outputs is same. In parsing checkpoint format, input and output needs to be provided.
For checkpoint version 2:
$ nnabla_cli convert input.ckpt.meta output.nnp --inputs x0,x1 --outputs y0,y1
In the same directory of input.ckpt.meta, the related files, such as checkpoint, *.ckpt.index, … and so on are required to exist.
Convert Tensorflow saved_model to NNP¶
$ nnabla_cli convert input_saved_model output.nnp
Convert NNP to Tensorflow Lite¶
$ nnabla_cli convert input.nnp output.tflite
Convert Tensorflow Lite to NNP¶
$ nnabla_cli convert input.tflite output.nnp
Splitting network¶
Splitting network is a bit complicated and can be troublesome.
NNP file could have multiple Executor networks, but Split supports only single network to split.
First, you must confirm how many Executors there are in the NNP, and specify what executor to split with nnabla_cli dump.
$ nnabla_cli dump squeezenet11.files/SqueezeNet-1.1/*.{nntxt,h5}
2018-08-27 15:02:40,006 [nnabla][INFO]: Initializing CPU extension...
Importing squeezenet11.files/SqueezeNet-1.1/net.nntxt
Importing squeezenet11.files/SqueezeNet-1.1/parameters.h5
Expanding Training.
Expanding Top5Error.
Expanding Top1Error.
Expanding Runtime.
Optimizer[0]: Optimizer
Optimizer[0]: (In) Data variable[0]: Name:TrainingInput Shape:[-1, 3, 480, 480]
Optimizer[0]: (In) Data variable[1]: Name:SoftmaxCrossEntropy_T Shape:[-1, 1]
Optimizer[0]: (Out)Loss variable[0]: Name:SoftmaxCrossEntropy Shape:[-1, 1]
Monitor [0]: train_error
Monitor [0]: (In) Data variable[0]: Name:Input Shape:[-1, 3, 320, 320]
Monitor [0]: (In) Data variable[1]: Name:Top5Error_T Shape:[-1, 1]
Monitor [0]: (Out)Monitor variable[0]: Name:Top5Error Shape:[-1, 1]
Monitor [1]: valid_error
Monitor [1]: (In) Data variable[0]: Name:Input Shape:[-1, 3, 320, 320]
Monitor [1]: (In) Data variable[1]: Name:Top1rror_T Shape:[-1, 1]
Monitor [1]: (Out)Monitor variable[0]: Name:Top1rror Shape:[-1, 1]
Executor [0]: Executor
Executor [0]: (In) Data variable[0]: Name:Input Shape:[-1, 3, 320, 320]
Executor [0]: (Out)Output variable[0]: Name:y' Shape:[-1, 1000]
As above output now you know only 1 executor.
Then you can show executor information with nnabla_cli dump -E0.
$ nnabla_cli dump -E0 squeezenet11.files/SqueezeNet-1.1/*.{nntxt,h5}
2018-08-27 15:03:26,547 [nnabla][INFO]: Initializing CPU extension...
Importing squeezenet11.files/SqueezeNet-1.1/net.nntxt
Importing squeezenet11.files/SqueezeNet-1.1/parameters.h5
Try to leave only executor[Executor].
Expanding Runtime.
Executor [0]: Executor
Executor [0]: (In) Data variable[0]: Name:Input Shape:[-1, 3, 320, 320]
Executor [0]: (Out)Output variable[0]: Name:y' Shape:[-1, 1000]
You can get list of function adding -F option.
$ nnabla_cli dump -FE0 squeezenet11.files/SqueezeNet-1.1/*.{nntxt,h5}
2018-08-27 15:04:10,954 [nnabla][INFO]: Initializing CPU extension...
Importing squeezenet11.files/SqueezeNet-1.1/net.nntxt
Importing squeezenet11.files/SqueezeNet-1.1/parameters.h5
Try to leave only executor[Executor].
Expanding Runtime.
Executor [0]: Executor
Executor [0]: (In) Data variable[0]: Name:Input Shape:[-1, 3, 320, 320]
Executor [0]: (Out)Output variable[0]: Name:y' Shape:[-1, 1000]
Executor [0]: Function[ 0 ]: Type: Slice Name: Slice
Executor [0]: Function[ 1 ]: Type: ImageAugmentation Name: ImageAugmentation
Executor [0]: Function[ 2 ]: Type: MulScalar Name: SqueezeNet/MulScalar
Executor [0]: Function[ 3 ]: Type: AddScalar Name: SqueezeNet/AddScalar
Executor [0]: Function[ 4 ]: Type: Convolution Name: SqueezeNet/Convolution
Executor [0]: Function[ 5 ]: Type: ReLU Name: SqueezeNet/ReLU
Executor [0]: Function[ 6 ]: Type: MaxPooling Name: SqueezeNet/MaxPooling
SNIP...
Executor [0]: Function[ 63 ]: Type: ReLU Name: SqueezeNet/FireModule_8/Expand1x1ReLU
Executor [0]: Function[ 64 ]: Type: Concatenate Name: SqueezeNet/FireModule_8/Concatenate
Executor [0]: Function[ 65 ]: Type: Dropout Name: SqueezeNet/Dropout
Executor [0]: Function[ 66 ]: Type: Convolution Name: SqueezeNet/Convolution_2
Executor [0]: Function[ 67 ]: Type: ReLU Name: SqueezeNet/ReLU_2
Executor [0]: Function[ 68 ]: Type: AveragePooling Name: SqueezeNet/AveragePooling
Executor [0]: Function[ 69 ]: Type: Reshape Name: SqueezeNet/Reshape
Executor [0]: Function[ 70 ]: Type: Identity Name: y'
If you want to get network without Image Augmentation, according to above output, ImageAugmentation is placed on index 2.
With splitting after index 3, you can get network without ImageAugmentation.
You must specify -E0 -S 3- option to nnabla_cli convert
This command rename output to XXX_S_E.nnp, XXX is original name, S is start function index, and E is end function index.
$ nnabla_cli convert -E0 -S 3- squeezenet11.files/SqueezeNet-1.1/*.{nntxt,h5} splitted.nnp
2018-08-27 15:20:21,950 [nnabla][INFO]: Initializing CPU extension...
Importing squeezenet11.files/SqueezeNet-1.1/net.nntxt
Importing squeezenet11.files/SqueezeNet-1.1/parameters.h5
Try to leave only executor[Executor].
Expanding Runtime.
Shrink 3 to 70.
Output to [splitted_3_70.nnp]
Finally you got splitted_3_70.nnp as splitted output.
You can check splitted NNP with nnabla_cli dump
NOTE: Input shape is changed from original network. New input shape is same as start function’s input.
$ nnabla_cli dump splitted_3_70.nnp
2018-08-27 15:20:28,021 [nnabla][INFO]: Initializing CPU extension...
Importing splitted_3_70.nnp
Expanding Runtime.
Executor [0]: Executor
Executor [0]: (In) Data variable[0]: Name:SqueezeNet/MulScalar Shape:[-1, 3, 227, 227]
Executor [0]: (Out)Output variable[0]: Name:y' Shape:[-1, 1000]
Done.